An integrated knowledge database dedicated to ncRNAs, especially lncRNAs.



An integrated knowledge database dedicated to ncRNAs, especially lncRNAs.



| What is NONCODE ? | |
| NONCODE (current version v5.0) is an integrated knowledge database dedicated to non-coding RNAs (excluding tRNAs and rRNAs). Now, there are 17 species in NONCODE(human, mouse, cow, rat, chicken, fruitfly, zebrafish, celegans, yeast, Arabidopsis, chimpanzee, gorilla, orangutan, rhesus macaque, opossum platypus and pig). The source of NONCODE includes literature and other public databases. We searched PubMed using key words ‘ncrna’, ‘noncoding’, ‘non-coding’,‘no code’, ‘non-code’, ‘lncrna’ or ‘lincrna. We retrieved the new identified lncRNAs and their annotation from the Supplementary Material or web site of these articles. Together with the newest data from Ensembl , RefSeq, lncRNAdb and GENCODE were processed through a standard pipeline for each species. The pipeline includes six steps:
Now, there are 17 species in NONCODE. All in all, NONCODE tries to present the most complete collection and annotation of non-coding RNA.It not only provides the basic information of lncRNA such as location, strand, exon number, length and sequence, but also the advanced information such as the expression profile, exosome expression profile, conservation info, predicted function and disease relation. The genome version of each species in current NONCODE version 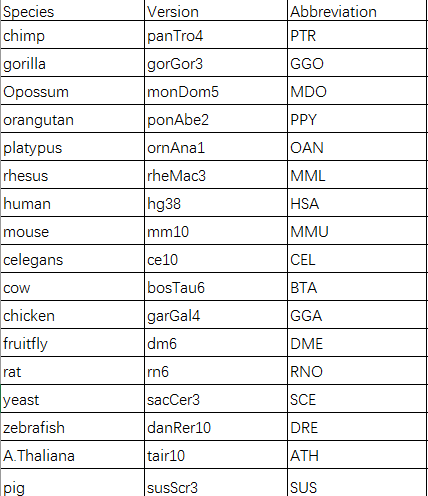
| |